Science
Deep Learning Transforms 3D Imaging of Fruit Tissue Structures
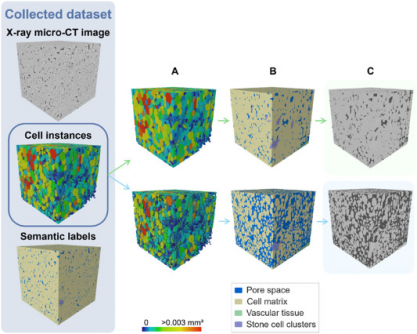
Researchers at KU Leuven have unveiled a groundbreaking deep learning model that accurately segments and quantifies the 3D microstructure of fruit tissues, specifically in apples and pears. Published in the journal Plant Phenomics on July 5, 2025, this novel approach significantly surpasses traditional 2D imaging methods and earlier algorithms in terms of precision and efficiency.
Conventional microscopy techniques for examining plant tissues require extensive preparation and often provide limited fields of view. While X-ray micro-computed tomography (micro-CT) has emerged as a promising non-destructive imaging tool, challenges remain in quantifying tissue morphology. Overlapping features and low image contrast complicate the analysis, hindering the accurate separation of crucial components such as parenchyma cells, vascular tissues, and stone cell clusters.
Recent advancements in deep learning have revolutionized image analysis in various fields, including medicine and biology. Recognizing the potential for plant research, the team led by Pieter Verboven developed an automated framework that leverages deep learning techniques for improved 3D segmentation of plant tissues from native X-ray micro-CT images.
Innovative Framework for Plant Tissue Analysis
The study introduced a sophisticated 3D panoptic segmentation framework. This framework incorporates the 3D extension of Cellpose alongside a 3D Residual U-Net architecture. As a result, it achieves comprehensive labeling of fruit tissue microstructures derived from X-ray micro-CT images. The model performs both instance and semantic segmentation, effectively predicting intermediate gradient fields across three dimensions to isolate individual parenchyma cells while classifying voxels into distinct categories such as cell matrix, pore space, vasculature, or stone cell clusters.
To enhance model training, the researchers employed datasets of apples and pears augmented with synthetic data. Techniques included morphological dilation and erosion, grey-value assignment, and the addition of Gaussian noise. Rigorous benchmarking against a 2D instance segmentation model and a marker-based watershed algorithm revealed that the new 3D model achieved remarkable performance. It registered Aggregated Jaccard Index (AJI) scores of 0.889 for apples and 0.773 for pears, outperforming the 2D model’s scores of 0.861 and 0.732, respectively.
Evaluation metrics, including the Dice Similarity Coefficient (DSC), confirmed the model’s efficacy in accurately segmenting tissue components. The segmentation of pore spaces and cell matrices approached perfection, while the model successfully identified vasculature with DSC scores of 0.506 in apples and 0.789 in pears. Additionally, it demonstrated strong performance in recognizing stone cell clusters, with scores reaching 0.683 for Intersection over Union (IoU) and 0.810 for DSC.
Implications for Plant Research
Visual validation corroborated the model’s accurate detection of vascular bundles in varieties such as ‘Kizuri’ and ‘Braeburn’ apples, alongside the smooth segmentation of stone cell clusters in ‘Celina’ and ‘Fred’ pears, achieving DSC scores of up to 0.90. Despite these successes, the study noted that additional data augmentation and targeted subsets did not enhance performance, likely due to dataset imbalance and domain shifts.
Morphometric analysis provided further validation, revealing vasculature widths ranging from 70 to 780 μm and variable dimensions and sphericity of stone cell clusters (averaging between 0.68 and 0.74). This deep learning model stands out as the most comprehensive automated solution to date for quantifying plant tissue microstructure, offering a non-destructive tool for plant scientists to investigate how microscopic structures affect water, gas, and nutrient transport.
The implications of this research extend beyond fruit analysis, providing a scalable framework applicable to various crops. The model enhances understanding of tissue development, ripening, and stress responses, potentially transforming practices in agriculture and food science. Its compatibility with standard X-ray micro-CT instruments positions it as an accessible solution for integrating artificial intelligence into the study of plant anatomy.
Funding for this research was provided by the Research Foundation – Flanders (FWO) under grant number S003421N as part of the SBO project FoodPhase, alongside support from KU Leuven through project C1 C14/22/076. The study represents a significant leap forward in the intersection of deep learning technology and plant science, facilitating faster, more precise studies that may ultimately enhance agricultural productivity and sustainability.
-

 Top Stories1 month ago
Top Stories1 month agoUrgent Update: Tom Aspinall’s Vision Deteriorates After UFC 321
-
 Health1 month ago
Health1 month agoMIT Scientists Uncover Surprising Genomic Loops During Cell Division
-

 Science4 weeks ago
Science4 weeks agoUniversity of Hawaiʻi Joins $25.6M AI Project to Enhance Disaster Monitoring
-

 Top Stories1 month ago
Top Stories1 month agoAI Disruption: AWS Faces Threat as Startups Shift Cloud Focus
-

 Science2 months ago
Science2 months agoTime Crystals Revolutionize Quantum Computing Potential
-

 World2 months ago
World2 months agoHoneywell Forecasts Record Business Jet Deliveries Over Next Decade
-

 Entertainment1 month ago
Entertainment1 month agoDiscover the Full Map of Pokémon Legends: Z-A’s Lumiose City
-

 Top Stories2 months ago
Top Stories2 months agoGOP Faces Backlash as Protests Surge Against Trump Policies
-

 Entertainment2 months ago
Entertainment2 months agoParenthood Set to Depart Hulu: What Fans Need to Know
-

 Politics2 months ago
Politics2 months agoJudge Signals Dismissal of Chelsea Housing Case Citing AI Flaws
-

 Sports2 months ago
Sports2 months agoYoshinobu Yamamoto Shines in Game 2, Leading Dodgers to Victory
-

 Health2 months ago
Health2 months agoMaine Insurers Cut Medicare Advantage Plans Amid Cost Pressures







