Science
Researchers Unlock RNA Folding with Advanced Simulation Techniques
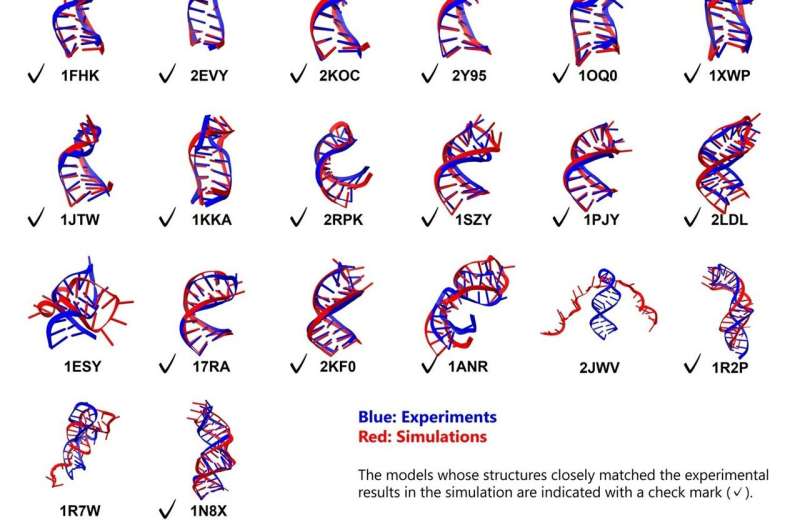
Groundbreaking research from the Tokyo University of Science has advanced understanding of RNA folding through enhanced molecular dynamics simulations. Published on November 4, 2025, in the journal ACS Omega, the study led by Associate Professor Tadashi Ando rigorously evaluated modern computational methods to simulate the folding processes of RNA stem-loops, significantly expanding the scope of previous research.
RNA plays a vital role in biological processes, extending beyond its function as a messenger of genetic information. Its ability to form complex three-dimensional structures is crucial for gene regulation and maintenance across living systems. As the demand for RNA-based therapeutics rises, accurately predicting these structures becomes essential for harnessing RNA’s full potential in biotechnology.
Historically, simulating RNA folding has proven challenging, with past studies achieving success only with simple stem-loop structures. The research team employed an innovative combination of the DESRES-RNA atomistic force field and the GB-neck2 implicit solvent model, which approximates the surrounding liquid as a continuous medium. This approach significantly improved the rate of conformational sampling over traditional explicit solvent models, which are more computationally intensive.
The study evaluated 26 RNA stem-loops, varying in length from 10 to 36 nucleotides, including structures with bulges and internal loops. All simulations commenced from fully extended, unfolded conformations. Remarkably, 23 out of 26 RNA molecules successfully folded into their anticipated structures, demonstrating the effectiveness of the chosen methodologies.
In the case of the 18 simpler stem loops, the simulations achieved exceptional accuracy, with root mean square deviation (RMSD) values—indicating deviation from known experimental structures—of less than 2 Å for the stems and less than 5 Å for the overall molecules. Notably, five out of eight more complex motifs, which included bulges or internal loops, also folded successfully, revealing distinct pathways for these structures.
Dr. Ando remarked on the significance of these findings, stating, “Previous studies have reported only two or three folding examples of simple stem-loop structures of approximately 10 residues, whereas this study deals with structures of much greater scale and complexity.” This research marks a pivotal milestone in computational biology, validating the reliability of the combined force field and solvent models for exploring large-scale conformational changes in RNA.
While the study achieved high accuracy for stem regions, the loop regions exhibited less precision, with RMSD values around 4 Å. This suggests that further optimization of the implicit solvent model parameters is necessary, particularly for modeling non-canonical base pairs and accounting for the effects of divalent cations like magnesium.
The implications of this research extend beyond theoretical modeling; understanding RNA folding is crucial for drug design targeting RNA molecules, which could lead to novel treatments for genetic disorders, viral infections such as COVID-19 and influenza, as well as certain cancers. Dr. Ando concluded, “The ability to reproduce the overall folding of the basic stem-loop structure is an important milestone in understanding and predicting the structure, dynamics, and function of RNA using highly accurate and reliable computational models. I expect this computational technique to lead to applications in RNA molecule design and drug discovery.”
This research represents a significant step forward in improving RNA-based medical tools and highlights the potential of advanced computational techniques in understanding complex biological systems.
-

 Top Stories1 month ago
Top Stories1 month agoUrgent Update: Tom Aspinall’s Vision Deteriorates After UFC 321
-
 Health1 month ago
Health1 month agoMIT Scientists Uncover Surprising Genomic Loops During Cell Division
-

 Science4 weeks ago
Science4 weeks agoUniversity of Hawaiʻi Joins $25.6M AI Project to Enhance Disaster Monitoring
-

 Top Stories1 month ago
Top Stories1 month agoAI Disruption: AWS Faces Threat as Startups Shift Cloud Focus
-

 Science2 months ago
Science2 months agoTime Crystals Revolutionize Quantum Computing Potential
-

 World2 months ago
World2 months agoHoneywell Forecasts Record Business Jet Deliveries Over Next Decade
-

 Entertainment1 month ago
Entertainment1 month agoDiscover the Full Map of Pokémon Legends: Z-A’s Lumiose City
-

 Top Stories2 months ago
Top Stories2 months agoGOP Faces Backlash as Protests Surge Against Trump Policies
-

 Entertainment2 months ago
Entertainment2 months agoParenthood Set to Depart Hulu: What Fans Need to Know
-

 Politics2 months ago
Politics2 months agoJudge Signals Dismissal of Chelsea Housing Case Citing AI Flaws
-

 Sports2 months ago
Sports2 months agoYoshinobu Yamamoto Shines in Game 2, Leading Dodgers to Victory
-

 Health2 months ago
Health2 months agoMaine Insurers Cut Medicare Advantage Plans Amid Cost Pressures







