Science
Researchers Launch SPRTA to Revolutionize Virus Evolution Tracking
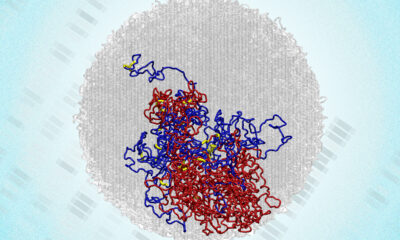
Researchers at the European Molecular Biology Laboratory (EMBL) and the Australian National University have introduced a groundbreaking tool called SPRTA (SPR-based Tree Assessment) to improve the evaluation of phylogenetic trees during viral outbreaks. This innovative method addresses the challenges posed by the vast amounts of genomic data generated during pandemics, enabling scientists to better understand virus evolution and transmission.
Enhancing Phylogenetic Analysis
Phylogenetic trees are essential for tracking the evolution and spread of viruses, such as SARS-CoV-2, which caused the global COVID-19 pandemic. Traditional methods, particularly the bootstrap method developed by Felsenstein in 1985, have long been used to measure confidence in these trees. However, this approach is inefficient when dealing with the millions of genomes sequenced during a pandemic.
The newly published paper in the journal Nature outlines how SPRTA provides a scalable, efficient alternative for evaluating the reliability of phylogenetic trees. It allows researchers to quickly identify the most trustworthy aspects of evolutionary relationships, significantly improving decision-making during outbreaks.
“For nearly 40 years, scientists have relied on the same method to measure confidence in evolutionary trees,” said Nick Goldman, Group Leader at EMBL-EBI. “But when faced with the scale of data we saw during the COVID-19 pandemic, the old method simply couldn’t cope.”
Innovative Approach to Reliability
Unlike traditional techniques that focus solely on the strength of sample groups, SPRTA evaluates the probability of a virus strain descending from a specific ancestor while considering alternative evolutionary paths. The tool tests various scenarios by virtually rearranging tree branches and assessing how well each configuration fits the available data.
SPRTA then assigns probability scores, providing a clear view of which relationships among strains are solid and where caution is warranted. “With SPRTA, we’re not just making phylogenetic tree-building faster, we’re making it smarter,” commented Nicola De Maio, Senior Scientist at EMBL-EBI.
This method has proven effective with over two million SARS-CoV-2 genomes, allowing researchers to highlight reliable areas within the phylogenetic tree, identify uncertain placements due to incomplete data, and propose credible alternative origins for certain branches.
SPRTA is integrated into MAPLE, a tool developed by EMBL-EBI for efficiently constructing large phylogenetic trees. It is also available in IQ-TREE, one of the most widely used software packages in phylogenetic analysis. This accessibility ensures that researchers worldwide can utilize SPRTA for outbreak tracking, genomic surveillance, and evolutionary studies.
In conclusion, the development of SPRTA represents a significant advancement in the ability to assess phylogenetic confidence at a pandemic scale. This tool not only enhances the speed and reliability of evolutionary analysis but also equips scientists with the necessary resources to respond effectively to future health crises.
-

 Top Stories1 month ago
Top Stories1 month agoUrgent Update: Tom Aspinall’s Vision Deteriorates After UFC 321
-
 Health1 month ago
Health1 month agoMIT Scientists Uncover Surprising Genomic Loops During Cell Division
-

 Science4 weeks ago
Science4 weeks agoUniversity of Hawaiʻi Joins $25.6M AI Project to Enhance Disaster Monitoring
-

 Top Stories1 month ago
Top Stories1 month agoAI Disruption: AWS Faces Threat as Startups Shift Cloud Focus
-

 Science2 months ago
Science2 months agoTime Crystals Revolutionize Quantum Computing Potential
-

 World2 months ago
World2 months agoHoneywell Forecasts Record Business Jet Deliveries Over Next Decade
-

 Entertainment1 month ago
Entertainment1 month agoDiscover the Full Map of Pokémon Legends: Z-A’s Lumiose City
-

 Top Stories2 months ago
Top Stories2 months agoGOP Faces Backlash as Protests Surge Against Trump Policies
-

 Entertainment2 months ago
Entertainment2 months agoParenthood Set to Depart Hulu: What Fans Need to Know
-

 Politics2 months ago
Politics2 months agoJudge Signals Dismissal of Chelsea Housing Case Citing AI Flaws
-

 Sports2 months ago
Sports2 months agoYoshinobu Yamamoto Shines in Game 2, Leading Dodgers to Victory
-

 Health2 months ago
Health2 months agoMaine Insurers Cut Medicare Advantage Plans Amid Cost Pressures








